What is Shine Dalgarno Sequence?
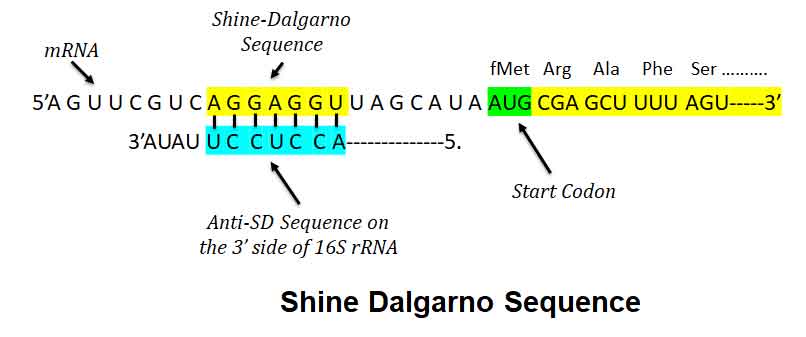
The Shine Dalgarno sequence aka SD Sequece is a ribosomal binding site in bacterial and archaeal mRNA. Usually, it is located around 8 bases upstream of the start codon (AUG). The SD sequence helps to recruit the ribosome to the mRNA to initiate protein synthesis by aligning the ribosome with the start codon. Once recruited, tRNA may add amino acids in sequence as dictated by the codons, moving downstream from the translational start site. In this article we will learn, what is Shine Dalgarno Sequence?
Genes are read in groups of three letters, but the ribosome needs to know where to start. For example, if you read “The big cat ate rat.”, it makes sense, but if you shift the starting point by one letter, it becomes “heb igc ata ter at_”. The SD sequence tells bacteria where to start protein synthesis so that the genes are read correctly.
Molecular Biology Notes | Molecular Biology PPTs | Molecular Biology MCQs
Ø The Shine Dalgarno sequence was proposed by Australian scientists John Shine and Lynn Dalgarno in 1973.
Ø The SD sequence is common in bacteria, but rarer in archaea.
Ø It is also present in some chloroplast and mitochondrial transcripts.
Ø The presence of SD sequence in chloroplast and mitochondrial transcript is considered as one of the strong pieces of evidence for endosymbiont theory.
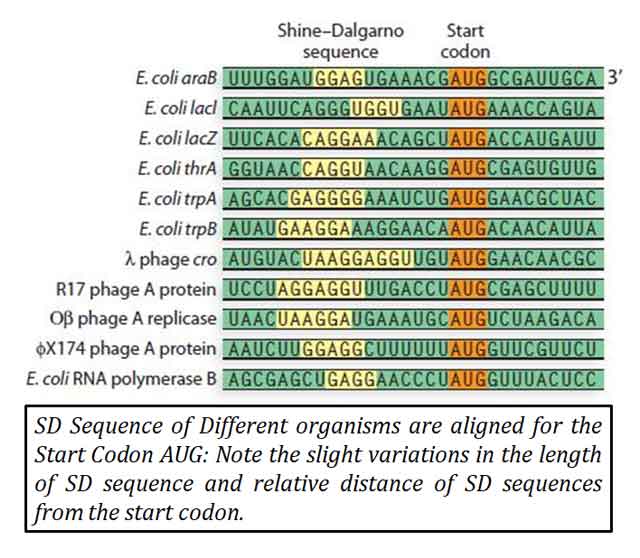
Ø The SD sequence typically consists of six base consensus sequences.
Ø Typically, the six-base consensus sequence is AGGAGG.
Ø In Escherichia coli, the SD sequence is AGGAGGU.
Ø The SD sequence, typically found upstream of the start codon on mRNA.
Ø The SD sequence is a part of Ribosome Binding Site (RBS*) in the mRNA.
Ø (* RBS: Ribosomal binding site is a sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of translation).
| You may also like NOTES in... | ||
|---|---|---|
| BOTANY | BIOCHEMISTRY | MOL. BIOLOGY |
| ZOOLOGY | MICROBIOLOGY | BIOSTATISTICS |
| ECOLOGY | IMMUNOLOGY | BIOTECHNOLOGY |
| GENETICS | EMBRYOLOGY | PHYSIOLOGY |
| EVOLUTION | BIOPHYSICS | BIOINFORMATICS |
Ø The nucleotide sequence SD sequence is complementary to a region of the 16S ribosomal RNA (rRNA) of the small ribosomal subunit (30S).
Ø The complementary sequence (CCUCCU) present in the 3’ end of 16S rRNA is called the Anti-Shine-Dalgarno sequence (ASD).
Ø Upon encountering the Shine-Dalgarno sequence, the ASD of the ribosome base pairs with it, after which translation is initiated.
Ø Thus, SD sequence acts as a binding site for the ribosome during the initiation phase of translation.
Ø This interaction facilitates the precise positioning of the ribosome on the mRNA molecule, ensuring accurate initiation of protein synthesis.
Functions of Shine Dalgarno Sequence
Ø The significance of the Shine Dalgarno sequence lies in its ability to promote efficient and precise translation initiation.
Ø By facilitating the proper alignment of the ribosome with the start codon on the mRNA, it ensures that translation begins at the correct site, thereby preventing errors in protein synthesis.
Ø SD sequence ensure the correct reading frame of codons in the mRNA.
Ø This precision is essential for the fidelity of gene expression and the production of functional proteins vital for cellular processes.
Ø SD sequence also helps in regulation of gene expression in prokaryotes (see below).
In summary, the Shine Dalgarno sequence represents a critical element in the machinery of translation initiation in prokaryotes. Its precise interaction with the ribosome ensures the accurate positioning of the translational machinery, leading to efficient and regulated protein synthesis.
FAQ on Shine Dalgarno Sequence
What will happen if the SD sequence is mutated?
Mutating an SD sequence can disrupt the ribosome’s ability to properly bind to the mRNA, leading to incorrect or inefficient initiation of protein synthesis. This can result in reduced or altered protein production, potentially affecting the organism’s function and viability.
Is the SD sequence is conserved among Prokaryotes?
Yes, the SD sequence is highly conserved among prokaryotes. It is a purine-rich sequence located upstream of the start codon in mRNA. The conservation of the SD sequence helps ensure efficient and accurate protein synthesis in prokaryotic organisms.
What is the similar component of Shine Dalgarno sequence in Eukaryotes?
In eukaryotes, the analogous component to SD sequence is the Kozak sequence. The Kozak sequence surrounds the start codon (AUG) and plays a critical role in the initiation of translation. It typically has the consensus sequence (GCC)GCCRCCAUGG, where R is a purine (adenine or guanine). The presence of the Kozak sequence helps the ribosome to identify the correct start codon for initiating protein synthesis.
How SD sequence helps in the regulation of gene expression?
SD sequence can also contribute to the regulation of gene expression by influencing the efficiency of translation initiation. Variations in the sequence or its distance from the start codon can modulate the rate of translation initiation, thereby impacting the abundance of specific proteins within the cell. This regulatory role allows organisms to fine-tune protein synthesis in response to changing environmental conditions or developmental cues.
What are the uses of SD sequences in biotechnology and Genetic engineering?
Shine Dalgarno (SD) sequences have several important uses in biotechnology and genetic engineering:
(1). Optimizing Protein Expression: By engineering SD sequences with optimal spacing and complementarity to the ribosomal RNA, scientists can enhance the translation efficiency and increase the yield of recombinant proteins in bacterial expression systems.
(2). Constructing Expression Vectors: SD sequences are included in plasmid vectors to ensure proper translation initiation of cloned genes. This is crucial for producing proteins in bacterial hosts such as E. coli.
(3). Gene Cloning and Synthetic Biology: SD sequences are used in designing synthetic gene constructs to ensure that the introduced genes are efficiently translated into proteins. This is important for applications like metabolic engineering and synthetic biology where precise control over protein production is required.
(4). Studying Gene Regulation: Researchers use modified SD sequences to study the effects of translation initiation on gene expression. By altering the SD sequence, scientists can investigate how changes in translation efficiency impact cellular processes.
What are the Functions of Shine Dalgarno sequence?
The Shine Dalgarno (SD) sequence is significant because it plays a critical role in the initiation of protein synthesis in prokaryotes. Key points of its significance include:
(1). Ribosome Binding: The SD sequence, is complementary to a sequence at the 3′ end of the 16S rRNA component of the small ribosomal subunit. This complementarity helps the ribosome to bind to the mRNA.
(2). Correct Start Codon Identification: By aligning the ribosome with the start codon (AUG), the SD sequence ensures that translation begins at the correct location on the mRNA.
(3). Translation Efficiency: A well-conserved SD sequence enhances the efficiency of translation initiation, leading to more effective and rapid protein synthesis.
(4). Gene Expression Regulation: Variations in the SD sequence can influence the strength of ribosome binding, thus regulating the level of gene expression. This allows cells to modulate protein production in response to different conditions.
| You may also like... | ||
|---|---|---|
| NOTES | QUESTION BANK | COMPETITIVE EXAMS. |
| PPTs | UNIVERSITY EXAMS | DIFFERENCE BETWEEN.. |
| MCQs | PLUS ONE BIOLOGY | NEWS & JOBS |
| MOCK TESTS | PLUS TWO BIOLOGY | PRACTICAL |

Pingback: The Central Dogma Part 2 – Microbiology Musings