Protein Structure
Learning objectives Structure of Proteins: Protein Structure: Primary Structure, Secondary Structure (Alpha Helix, Beta Plates, Beta Turns), Tertiary Structure, Quaternary Structure, Bonds Stabilizing different Protein Structures.
Protein Structure
Ø Proteins are the polymers of amino acids. Individual amino acids (residues) are joined by peptide bonds to form the linear polypeptide chain. This linear polypeptide chain is folded into specific structural conformations or simply ‘structure’. A protein can have up to four levels of structural conformations. Previously we have discussed but the ‘Bonds involved in Protein Structure”. In the present post, we will discuss different types of protein structures.
Ø A protein can have Four levels of structural organization:
Ø They are designated as:
(1). Primary Structure
(2). Secondary Structure
(3). Tertiary Structure
(4). Quaternary Structure
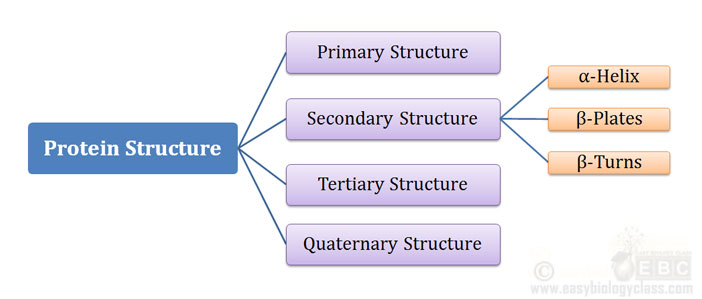
Ø Secondary, Tertiary and Quaternary structure are together called the three-dimensional (3D) structure of the protein.
Ø All functional proteins will have up to 3 (tertiary level) of structures.
Ø Some proteins will have all the 4 levels of structures (up to quaternary structure).
(1). Primary Structure
Ø Primary structure of a protein gives the details of the amino acid sequence of a protein.
Ø The primary structure will tell you two main things: (i) The number of amino acid residues in the protein and (ii) the sequence of amino acids.
| You may also like NOTES in... | ||
|---|---|---|
| BOTANY | BIOCHEMISTRY | MOL. BIOLOGY |
| ZOOLOGY | MICROBIOLOGY | BIOSTATISTICS |
| ECOLOGY | IMMUNOLOGY | BIOTECHNOLOGY |
| GENETICS | EMBRYOLOGY | PHYSIOLOGY |
| EVOLUTION | BIOPHYSICS | BIOINFORMATICS |
Ø The ‘sequence’ information contains the correct order of amino acids in the protein starting from N-terminal to C-terminal.
Ø The primary structure of a protein will determine all other levels of structural organization of a protein (secondary, tertiary and quaternary).
Ø The primary structure is stabilized by Peptide Bonds (Covalent Bond).
Ø The first study about an unknown protein will be its sequence determination (determination of primary structure).
Ø First sequenced protein: Insulin by Frederick Sanger.
Primary Structure of Insulin

Importance of Primary Structure:
Ø The primary structure of a protein will offer insights into its:
(a). Three dimensional (3D) structure
(b). Function of the protein
(c). Cellular location
(d). Evolution of the protein
Ø Primary structure data can be used for the sequence searching from the protein databases.
Three Dimensional Structures of Proteins
Ø The backbone of a protein contains hundreds of individual bonds.
Ø Free rotation is possible around many of these bonds.
Ø The free rotation allows an unlimited number of conformations around these bonds.
Ø However, each protein has a specific (unique) structural conformation.
Ø This unique structural formation of a protein is called its 3D structure.
Ø The spatial arrangement of atoms in a protein is called its ‘Conformation’.
Ø Proteins in their functional, folded conformations are called native proteins.
Ø The conformations of a protein are mainly stabilized by weak interactions such as hydrogen bonds, hydrophilic interactions, hydrophobic interactions etc..
Ø These weak interactions can be easy distorted with less expenditure of energy.
Ø Proteins may have three levels of Three Dimensional (3D) Organizations. They are:
Secondary structure
Tertiary structure
Quaternary structure
(2). Secondary Structure
Ø Secondary structure is the special local conformation of some part of a polypeptide chain.
Ø It is the folding pattern of the regular polypeptide backbone.
Ø Different types of secondary structures occur in nature.
Ø The secondary structures are stabilized mainly by Hydrogen Bonds.
Ø Three most important secondary structure in protein are:
A. α-Helix
B. β-Conformations (β-plates)
C. β-Turns
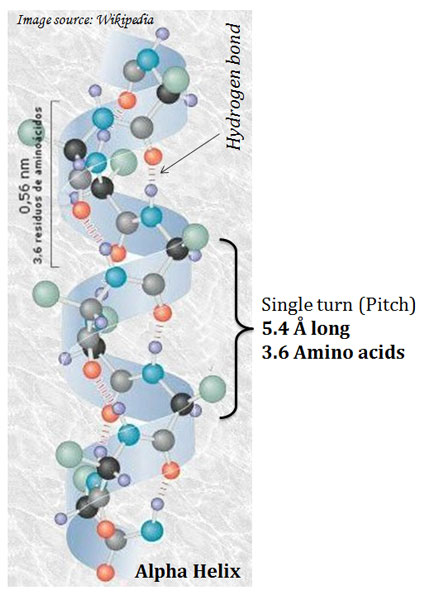
image source: wikipedia
(A). α-Helix
Ø The α-helix is the most common secondary structure.
Ø They are regular structures that repeat every 5.4 Å.
Ø It is the simplest arrangement of a polypeptide chain.
Ø The α-helical structure of the protein was proposed by Pauling and Corey in 1951.
Ø The polypeptide backbone is tightly wound around an imaginary axis drawn longitudinally through the middle of the helix, and the R groups of the amino acid residues protrude outward from the helical backbone.
Ø Pitch of helix: The repeating unit of the helix.
Ø The pitch is a single turn of the helix which extends about 5.4 Å.
Ø Each helical turn in α-helix contains 3.6 amino acids.
Ø The helical twist of the α-helix in all protein is right-handed.
Ø The α- helix is stabilized by hydrogen bonds.
Ø The α-helix is so common in protein because it makes optimal use of internal hydrogen bonds.
Ø Hydrogen bonds are formed between the hydrogen attached to the electronegative nitrogen atom of the peptide linkage and the electronegative carbonyl oxygen atom of the fourth amino acid on the amino-terminal side of the peptide bond.
Ø Within the α-helix, every peptide bond participates in hydrogen bonding.
Ø All hydrogen bonds together provide considerable stability to the α-helix.
Ø All polypeptide cannot form a sable α-helix.
Ø The interactions between the amino acid side chains can stabilize or destabilize the α-helix.
Ø For example, if a polypeptide chain has a long stretch of Glutamic Acid residues, this segment the chain will not form an α-helix.
Ø The negatively charged carboxyl group of the adjacent Glu residues repels each other strongly so that they prevent the formation of the α-helix.
Ø Similarly, a polypeptide rich in Proline will not form an α-helix.
Ø In proline, the nitrogen atom is part of a rigid ring and rotation about the N – Cα bond is NOT possible.
Ø Thus proline introduces a destabilizing kink in the polypeptide and hence proline is very rarely found in α-helix.

image source: wikipedia
β-Conformations (β-Plates)
Ø The β conformation or β-plates organize the polypeptide chains into sheets.
Ø The β-conformation is an extended form of a polypeptide chain.
Ø Here the polypeptide backbone is extended into a zigzag structure.
Ø The zigzag polypeptide chains can be arranged side-by-side to form a structure resembling a series of pleats called β-sheets.
Ø Here also the structure is stabilized by hydrogen bonds.
Ø However, unlike α-helix, the hydrogen bonds are formed between adjacent segments of the chain.
Ø The R-groups of adjacent amino acids protrude from the zigzag structure in opposite direction creating the alternating pattern.
Ø The polypeptide chains in the β-sheets may be arranged either in parallel (the same direction) or anti-parallel (opposite direction).
Ø Based on this the β-Plates are classified into two types: (diagram)
(a) Anti-parallel β-Plates
(b) Parallel β-Plates
(C). β-Turns:
Ø The β-turns are very common in proteins, where the peptide make a turn or loop (peptide make a reverse direction).
Ø In globular proteins, nearly one-third of the amino acid residues are in β turns.
Ø The β-turns are the connecting elements that link successive runs polypeptide chain.
Ø The β-turn connects the ends of two adjacent segments of anti-parallel β-sheets.
Ø The β-turn structure is an 180º turn involving four amino acid residues
Ø The carbonyl oxygen of the first residue forms a hydrogen bond with the amino group hydrogen of the fourth amino acid in the turn.
Ø Glycine (Gly) and Proline (Pro) resides frequently occurs in β-turns.
Ø Glycine due to its very small size (the R group is – H) allows β turns.
Ø Proline is an imino acid with its side chain covalently linked with the amino group.
Ø Proline residue in the peptide bond assumes the ‘cis’ configuration.
Ø The ‘cis’ conformation is very amenable to tight turns.
Ø There are several types of β turns of with type I and Type II are most common.
Ø The type I β-turn is found more than twice the frequency of type II.
Ø In type II, the third residue will always be a Glycine residue.
(3). Tertiary Structure Proteins
Ø Tertiary structure: The overall three-dimensional arrangement of all atoms in a protein is referred to as the tertiary structure.
Ø The tertiary structure will have a single polypeptide “backbone” with one or more secondary structures.
Ø Tertiary structure is defined by the atomic coordinates.
Ø Tertiary structures in a protein are stabilized by both covalent and non-covalent bonds.
Ø Covalent bond: Disulfide bonds (between two Cys residues)
Ø Non-covalent interactions: Ionic interactions (electrostatic attractions), hydrophilic interactions, van der Waals interactions.
Ø The term ‘Domain’ is used to denote a single functional unit of a protein.
Ø A protein may have many domains with specific functions.
Ø A protein with a single subunit only has up to the tertiary structure.

image source: wikipedia
(4). Quaternary Structure
Ø Majority of functional protein contains more than one polypeptide chains and such a protein is said to be oligomeric.
Ø Each peptide forms a sub-unit or monomer or protomer.
Ø Quaternary structure: The arrangement of protein monomers in three-dimensional complexes in a multi-subunit protein is called quaternary structure.
Ø For a protein to have a quaternary structure, it should fulfil two conditions:
§ It should have more than one polypeptide subunits
§ There should not have permanent (covalent) interaction between the subunits (like disulfide bond).
Ø Insulin does not have the quaternary structure even if it contains two subunits.
Ø The two polypeptides in insulin are covalently connected with two disulfide bonds.
Ø Thus, insulin can have up to tertiary structure (not quaternary structure).
Ø Bonds stabilizing quaternary structure: hydrogen bonds, hydrophilic interactions, hydrophobic interactions, van der Waals interactions.
| You may also like... | ||
|---|---|---|
| NOTES | QUESTION BANK | COMPETITIVE EXAMS. |
| PPTs | UNIVERSITY EXAMS | DIFFERENCE BETWEEN.. |
| MCQs | PLUS ONE BIOLOGY | NEWS & JOBS |
| MOCK TESTS | PLUS TWO BIOLOGY | PRACTICAL |
Overview:

Image Source: Wikipedia
Reference
Nelson, D.L., Lehninger, A.L. and Cox, M.M., 2008. Lehninger’s Principles of Biochemistry. Macmillan.
Do you have any Queries?
Please leave me in the Comments Section below.
I will be Happy to Read your Comments and Reply.
You might also like…
@. Protein Structure PPT (Download)
@. Bonds involved in Protein Structure
@. Classification of Proteins based on Structure and Functions

Can we download any app regarding easy bio class????